Ploomber: Maintainable and Collaborative Pipelines in Jupyter
Ploomber is an open-source framework that allows teams to develop maintainable pipelines in Jupyter.

Jupyter is a fantastic tool for data exploration. The ability to transform our data interactively and get immediate visual feedback allows us to understand it quickly. However, when working on large projects, collaboration can become difficult. Features such as live collaboration are a gigantic leap forward for teamwork. Still, it has to complement an asynchronous workflow that allows team members to work at different times, especially in a remote-first workplace.
Back in 2020, I introduced Ploomber at JupyterCon to help practitioners build maintainable and reproducible data workflows. Fortunately, the community is growing. We’ve received great feedback from teams that use Ploomber to develop production-ready pipelines using Jupyter, debunking the notion that notebooks are only for prototyping.
As we’ve gathered more feedback from our community, we realized that while users had improved the reproducibility of their workflows, team dynamics didn’t change much. In many cases, collaborators worked in isolation, sharing processed data, which severely hindered reproducibility.

Since its first release, Ploomber aimed to promote software development best practices to produce more maintainable data projects. We’re now doubling our efforts to enable a collaborative and asynchronous workflow.
Enabling Code Reviews
We frequently hear from teams using Jupyter notebooks that it’s challenging to manage .ipynb files. For example, we heard from a data scientist that his company considered banning Jupyter notebooks because they couldn't figure out how to follow software engineering best practices. Our fellow data scientist was highly frustrated by this situation since Jupyter turbocharges his ability to explore and understand data.
Managing .ipynb files is challenging for multiple reasons. Suppose I push a notebook to a git repository. Then, I add a comment and push it to the repository again. The difference between the two versions looks like this:

.ipynb files contain input and outputs in a single file, making them extremely useful when we want to share code and results with a colleague but complicates code reviews where we want to compare the previous version with the current one. However, we can fix this problem by changing the underlying file format.
Many practitioners don’t know that Jupyter is agnostic to the underlying file representation, allowing us to interact with different file formats as notebooks. Jupytext is a fantastic project that enables us to open various file formats such as .py and .md as notebooks.
Ploomber integrates with jupytext, allowing users to store their source code as .py files and explore data interactively with Jupyter. Since source code exists in .py files, this enables code reviews, file merging, and the flexibility to edit the code either in Jupyter or in any text editor, giving anyone on the team the freedom to use whatever tool they like the most.

Enabling Modularization
Often, teams develop significant parts of a project in a single notebook for convenience; however, this makes it hard to maintain the project in the long run. We know from decades of advancement in software engineering practice that modularizing our code produces more maintainable projects. Yet, we tend to work on single notebooks because breaking down analysis in multiple parts involves managing project structure, ensuring we route outputs correctly, and writing code to orchestrate all steps.
Ploomber allows users to concatenate multiple notebooks into a pipeline in two steps: list the notebooks in a YAML file and declare execution dependencies (e.g., download data, then clean it). Furthermore, Ploomber parses our execution dependencies and injects inputs into our notebook when opening it. Thus, there’s no need to hard-code any paths. The following image illustrates how to declare a pipeline in a pipeline.yaml file and the notebook's code injection process:
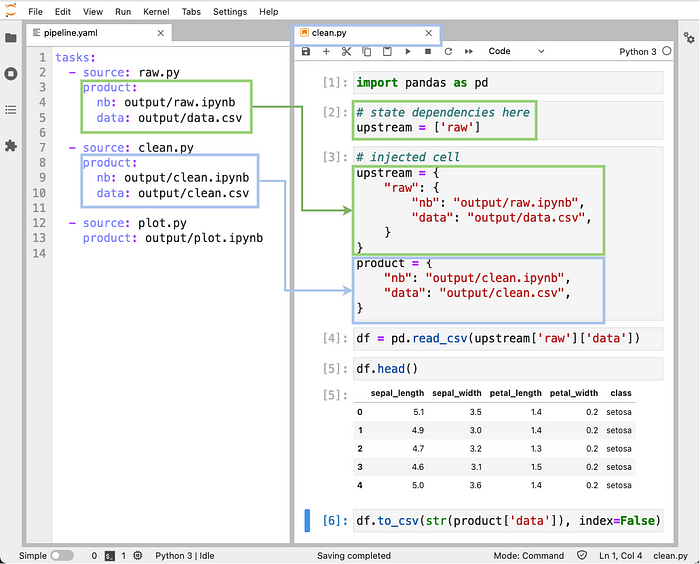
Modularization has another benefit. It allows teams to collaborate on separate, well-defined streams of work. Data projects have a sequential nature, and by explicitly structuring them into small tasks, it is easy to assign parts to various team collaborators. For example, if a two-person team is working on a project that uses two sources of data, each member can take one dataset:
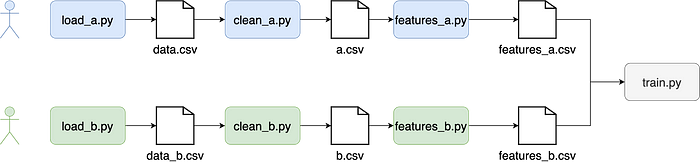
Enabling Testing
Modularization facilitates testing. A recommended practice when developing data pipelines is to test the output data from each task to ensure it has some minimum quality. Since we have clear boundaries among tasks, we can embed integration tests that check the output coming out of each step, ensuring that we know when something breaks.

Examples of integration tests include: checking there are no NULL values in a specific column or verifying values fall into a certain range. In Ploomber, you can execute an arbitrary function after your notebook finishes to assert statements on your data:

Ensuring Reproducibility
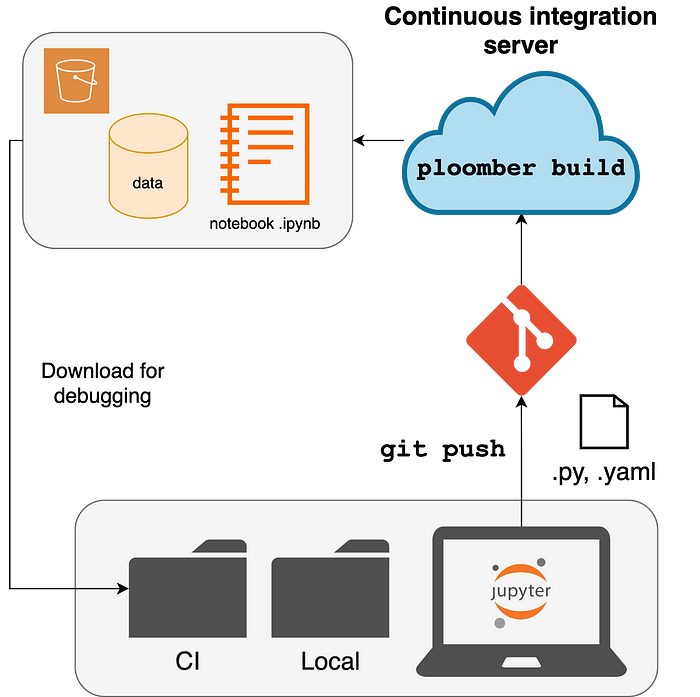
One of Jupyter notebook’s most recurring problems is hidden state; this happens when we execute code cells in an arbitrary order. However, when running cells in sequential order, the results do not match the recorded output.
Hidden state creates significant issues. For example, teams often execute notebooks locally, store them in a git repository, and don’t execute them again. For instance, I heard from a fellow data scientist that her team struggled to update a model in production because of a broken notebook that prevented her team from re-training the model: the notebook had been executed locally once but never tested for reproducibility.
The answer to hidden state is straightforward: test continuously. In software engineering, it’s common to run code and test it on each git push. However, such practice hasn't found its way into the data world, primarily because running data processing code may take hours, making testing unfeasible.
Fortunately, most errors are detectable with small amounts of data, and Ploomber simplifies managing pipeline configurations. For example, a user can define a sample parameter to run the pipeline with a fraction of the input data:

Then, a continuous integration script can run the pipeline with a sample by switching the sample parameter:
Furthermore, users can easily download remote artifacts to debug broken pipelines.
Enabling Pull Requests
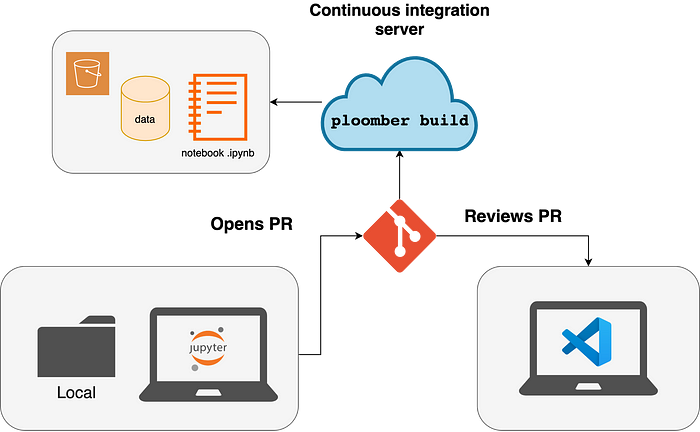
Once a new feature is ready, a team member can open a pull request. Since the code is in .py files, the reviewer can easily compare code versions. However, metrics or charts are essential to evaluate data processing code. For this reason, Ploomber generates a .ipynb file for each .py file; such executed notebooks can be attached to a pull request to review code and output.
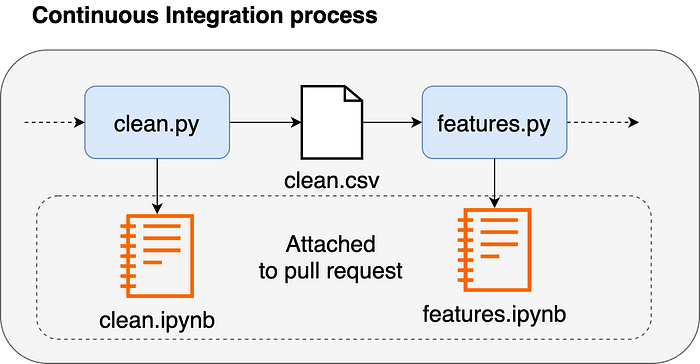
A CI process can orchestrate pipeline execution with a single command: ploomber build. However, if working with large datasets, Ploomber can export pipelines to execute in AWS Batch, Airflow, or Kubernetes (Argo Workflows).
Enabling Fast Iterations
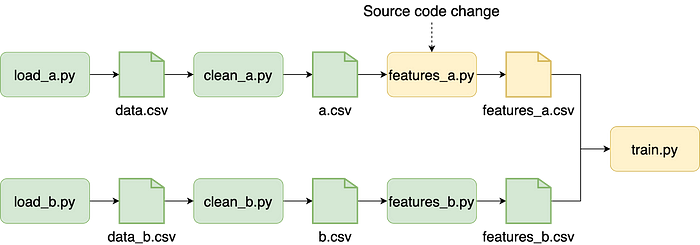
Data projects are highly iterative and require us to run small experiments to evaluate our final results. For example, we may add a new feature and assess whether that improves model performance. The more experiments we try, the higher our chance of success.
Such experiments are small and often only touch a small portion of the pipeline. If most of our tasks are unaffected, re-running tasks is a waste of time since they’ll generate the same results. Given that a training pipeline may take hours to run, it is essential to speed things up as much as possible. To enable faster iterations, Ploomber builds pipelines incrementally, skipping tasks whose source code hasn’t changed. Incremental builds help in multiple scenarios, for example, when trying out local experiments or running the pipeline in the CI system. Furthermore, they enable crash recovery: if we execute our pipeline and it crashes, we can fix the failing task, submit it again, and execution will take off from the point of failure.
JupyterLab Is a Production-Ready Platform
It is common for teams to develop prototypes in Jupyter, then refactor them into Python modules; such an approach creates a lot of overhead and a tremendous burden for data scientists (who produce the models) and engineers (who have to refactor notebook-based prototypes). Instead, we believe data projects should start with production in mind by following best software development practices that allow teams to iterate quickly.
Providing such an experience is challenging. Nevertheless, we are working hard to achieve that goal; we want data scientists and engineers to collaborate to produce production-ready projects that instantly go from Jupyter to production.
The Future
There is still a long way to capture our vision of the future of data workflows. Furthermore, we want to keep building the project with the help of our users. So if you’re interested in building Ploomber with us, join our community, or follow us on Twitter! And if you believe in our mission, please show your support with a star on GitHub.
Thanks to Ido Michael and Filip Jankovic for providing feedback.

